Synspace
Only lead discovery software exploring synthetically feasible chemical space
Created for all medicinal and computational chemists
Key Design Modules
- Two automated de novo generative molecular design methods
- Two scaffold hopping methods
- Automated fragment or lead growing in protein pockets
- Automated lead growing at selected positions
- Multi-step focused library enumeration
- Retrosynthesis
Features
- Embedded IP and availability checks
- All products have schemes & reagent data
- User friendly: no cheminformatic or synthetic skills required
- All molecules possess synthetic routes and reagent data
- Macrocyclic design capability

1. Autogrow module setup page for automated pocket filling
Protein pdb file and bound ligand are required as input data. Boundaries of the sampled chemical space and the reagent sources are automatically defined by SynSpace but customization is allowed. Can be run from GUI or via API.
1. De novo generative design module setup page for automated optimization
The unique Derivatization design technology only requires the active ligand structure and optionally the user-selected structural areas for optimization. The method exhaustively samples synthesizable chemical space of relevant molecular features. Depending on the settings, it can be applied for exploration or exploitation in lead optimization. Can be run from GUI or via API.
2. De novo generative design result page including three key molecules with further improved binding activity
The generative module returns an exhaustive sampling of relevant synthesizable molecules that have the potantial to significantly improve – often multiple MPO – parameters included in the target product profile. Macrocyclic generative design capability is included. All in silico molecules are rapidly actionable with synthesis, reagent, and vendor data.
1. 1-Click scaffold design module setup page for scaffold hopping
Only the ligand structure is required as input data. The module evaluates synthetically acecssible ring systems that match the 3D shape and user-selected properties (aromaticity, H-bonding) of the input scaffold. Can be run from GUI or via API.
2. 1-Click scaffold design result page including a key novel scaffold for lead optimization
Module returns full molecules based on new scaffold replacement suggestions that have good shape and feature complementarity with the input structure. Synthetic feasibility of molecules containing new scaffolds is classified into easy and hard subsets with an option to see hard to synthesize rings as well. Synthetic schemes are optionally included.
1. Library enumeration module setup page for focused library generation
The most advanced library enumeration tool available. Synthetic sequences of up to 9 steps and multiple reactions in each step can be explored. Easy drag-and-drop interface. Simple reagent search options for customizing each step. Physicochemical property, cost and availability, or structural filters are built in. Can be run from GUI only.
2. Library enumeration result page including three key analogs included in the final patent
Module returns a synthesizable focused library with all molecules that satisfy the enumeration criteria. Multiple positions can be varied in a single job. Synthesis cost and reagent availability can be used to rank close analogs.
2. Retrosynthesis result page including multiple molecules with synthetic solutions
Module returns all molecules classified into solved and unsolved subsets. Solved molecules can contain up to 100,000 different routes with reagent cost and availability data as well as different ranking columns to assist selection of the best route(s) for synthesis or for focused library generation.
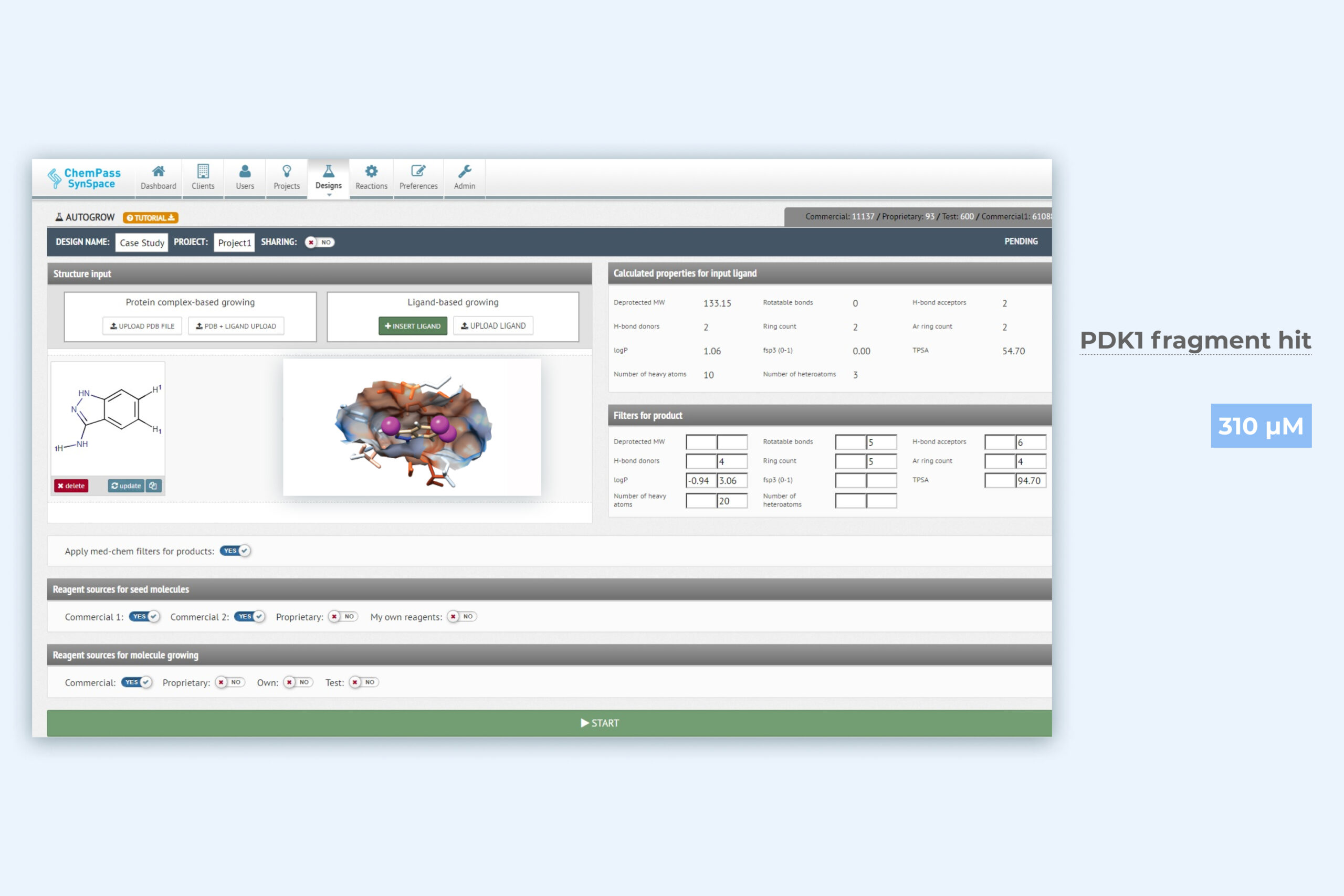
1. Autogrow module setup page for automated pocket filling
Protein pdb file and bound ligand are required as input data. Boundaries of the sampled chemical space and the reagent sources are automatically defined by SynSpace but customization is allowed. Can be run from GUI or via API.
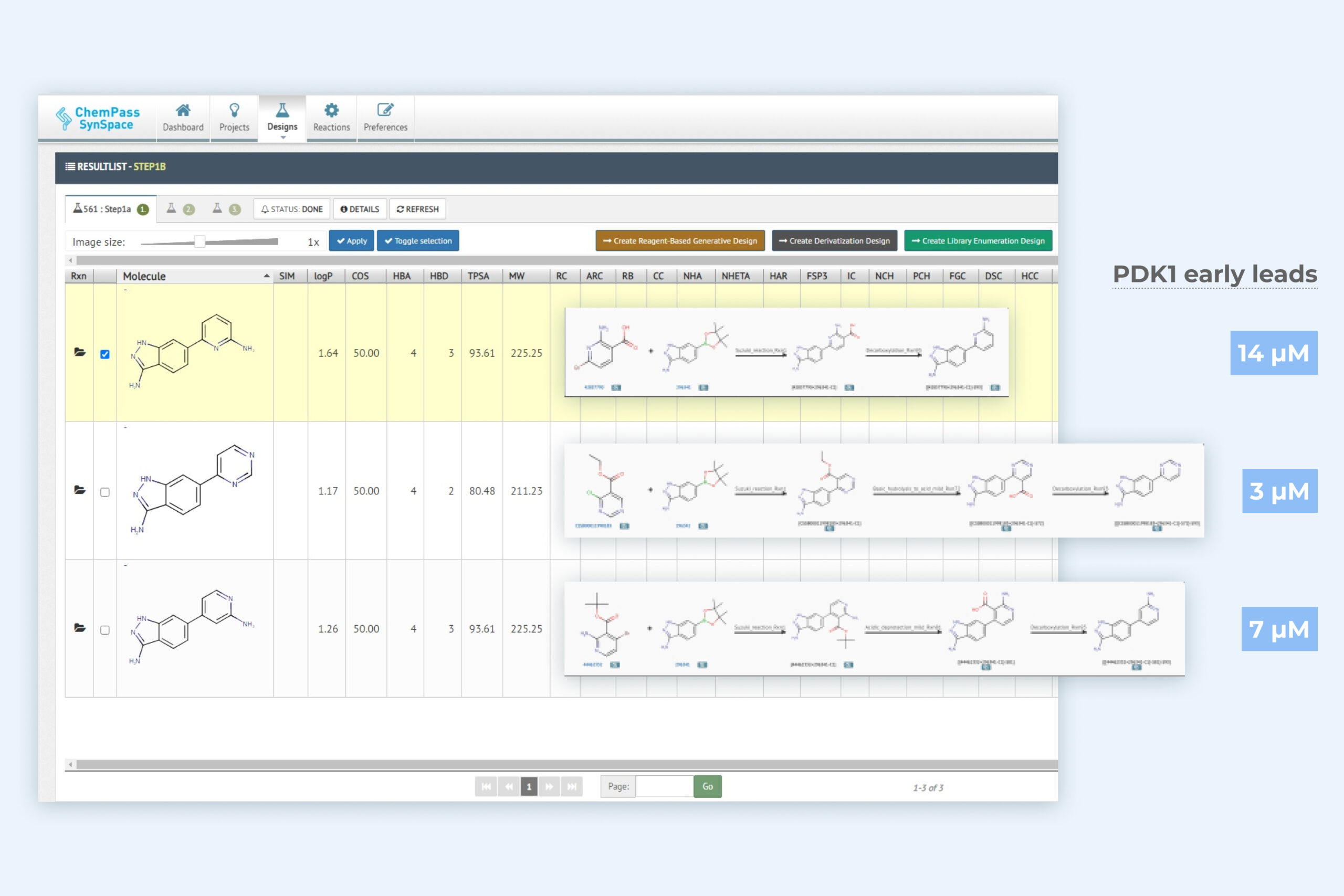
2. Autogrow result page including three key molecules with improved binding activity
Module returns synthesizable molecules that sample the chemical space in the direction(s) of the binding pocket where holes can be filled.
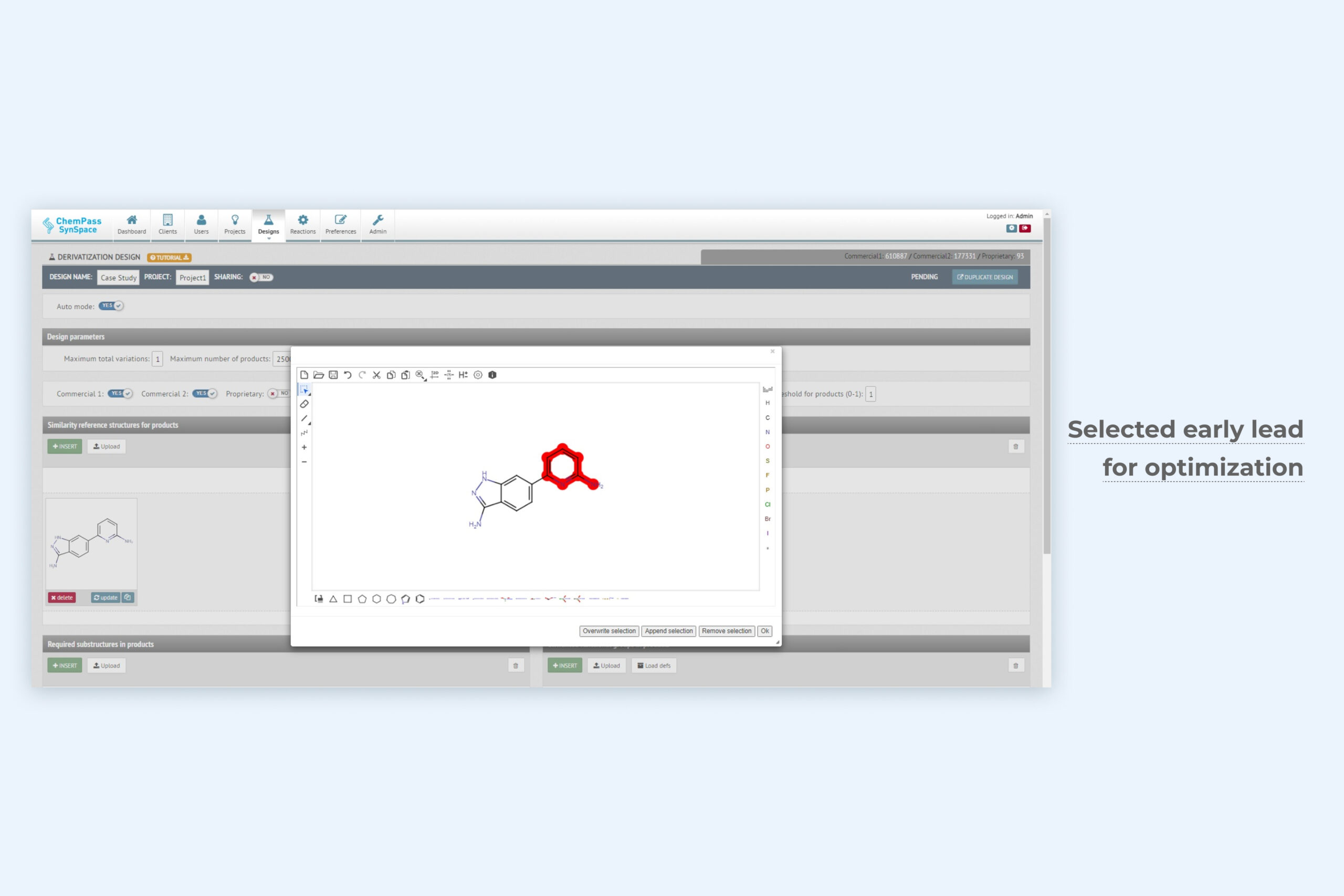
1. De novo generative design module setup page for automated optimization
The unique Derivatization design technology only requires the active ligand structure and optionally the user-selected structural areas for optimization. The method exhaustively samples synthesizable chemical space of relevant molecular features. Depending on the settings, it can be applied for exploration or exploitation in lead optimization. Can be run from GUI or via API.
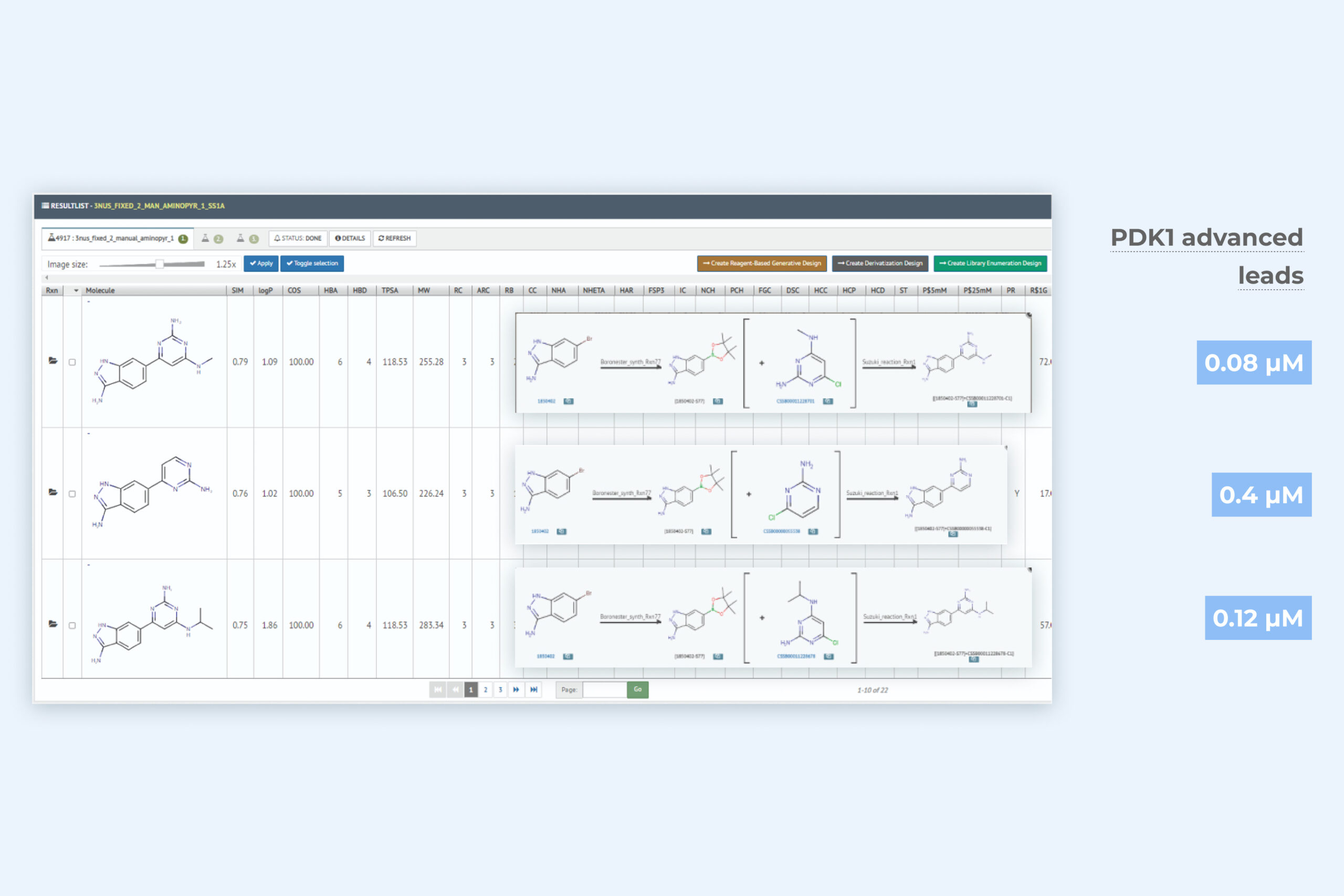
2. De novo generative design result page including three key molecules with further improved binding activity
The generative module returns an exhaustive sampling of relevant synthesizable molecules that have the potantial to significantly improve – often multiple MPO – parameters included in the target product profile. Macrocyclic generative design capability is included. All in silico molecules are rapidly actionable with synthesis, reagent, and vendor data.
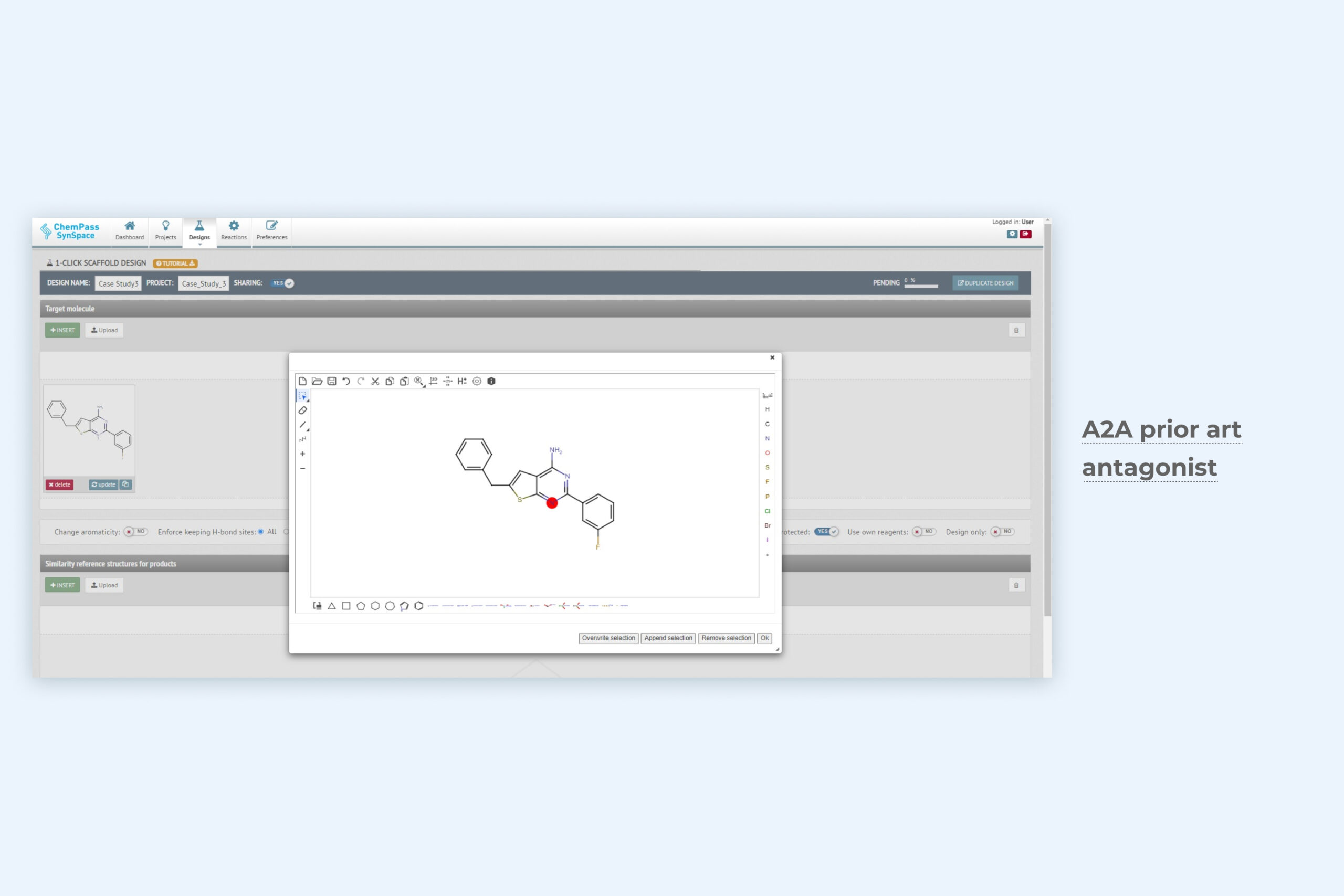
1. 1-Click scaffold design module setup page for scaffold hopping
Only the ligand structure is required as input data. The module evaluates synthetically acecssible ring systems that match the 3D shape and user-selected properties (aromaticity, H-bonding) of the input scaffold. Can be run from GUI or via API.
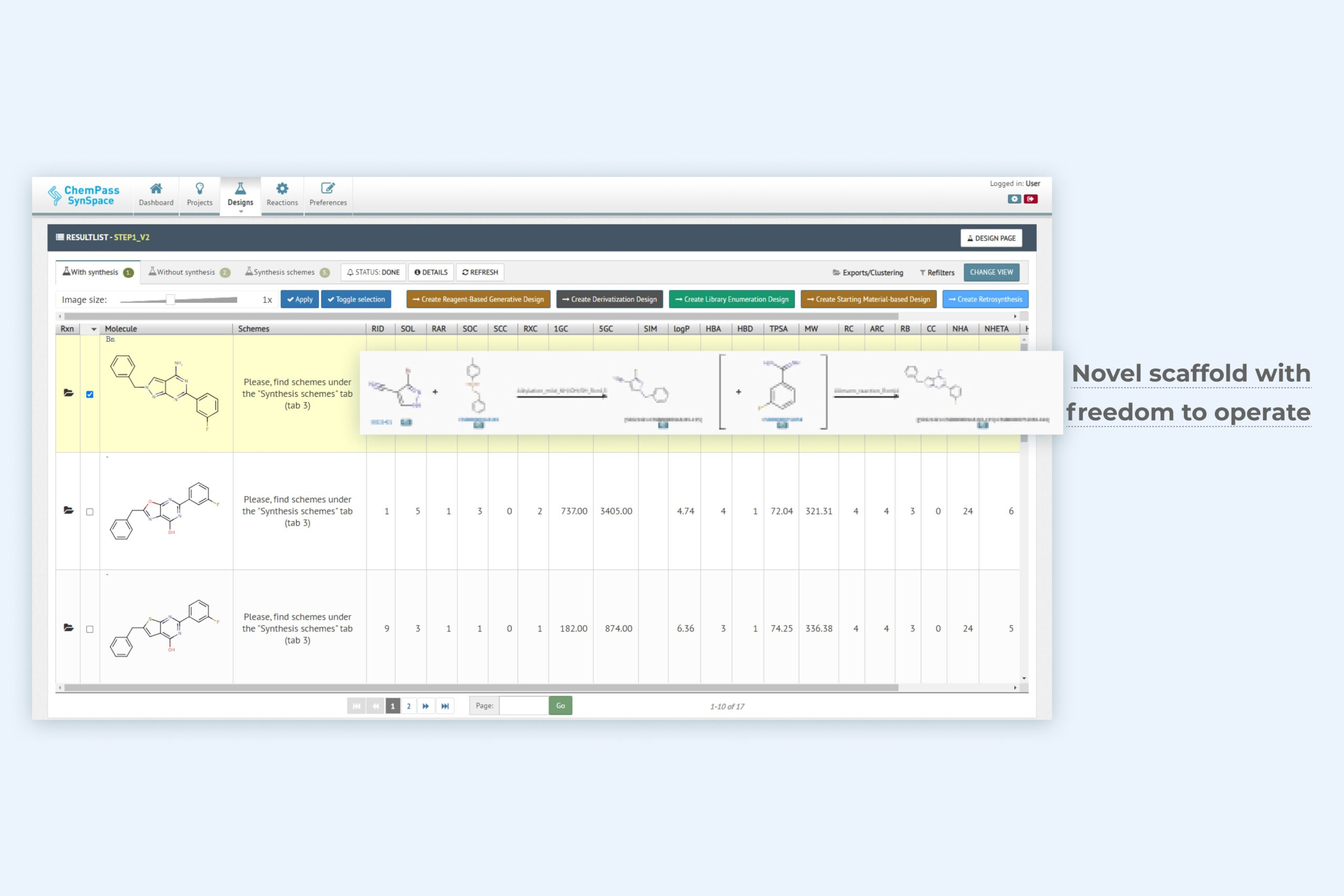
2. 1-Click scaffold design result page including a key novel scaffold for lead optimization
Module returns full molecules based on new scaffold replacement suggestions that have good shape and feature complementarity with the input structure. Synthetic feasibility of molecules containing new scaffolds is classified into easy and hard subsets with an option to see hard to synthesize rings as well. Synthetic schemes are optionally included.
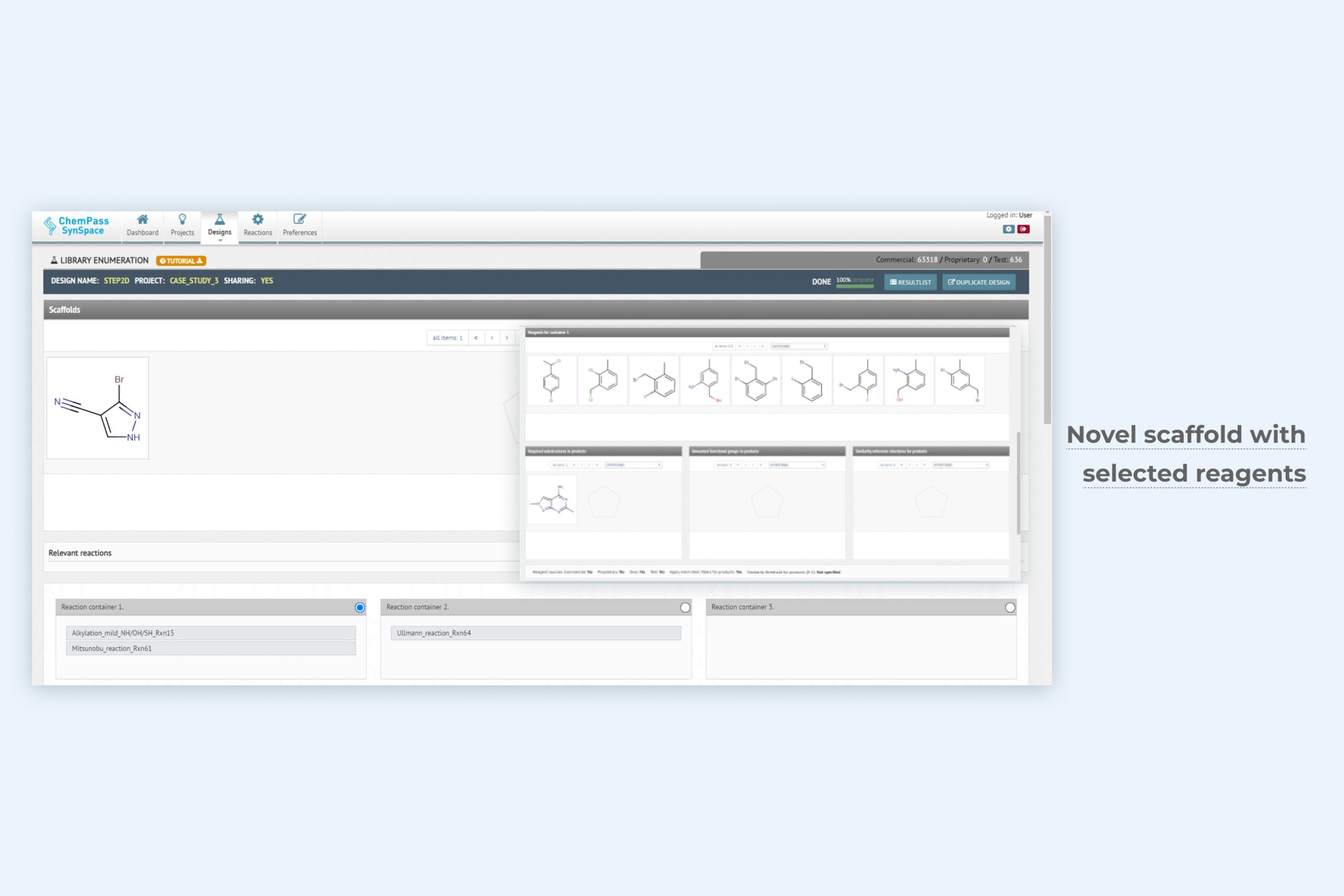
1. Library enumeration module setup page for focused library generation
The most advanced library enumeration tool available. Synthetic sequences of up to 9 steps and multiple reactions in each step can be explored. Easy drag-and-drop interface. Simple reagent search options for customizing each step. Physicochemical property, cost and availability, or structural filters are built in. Can be run from GUI only.
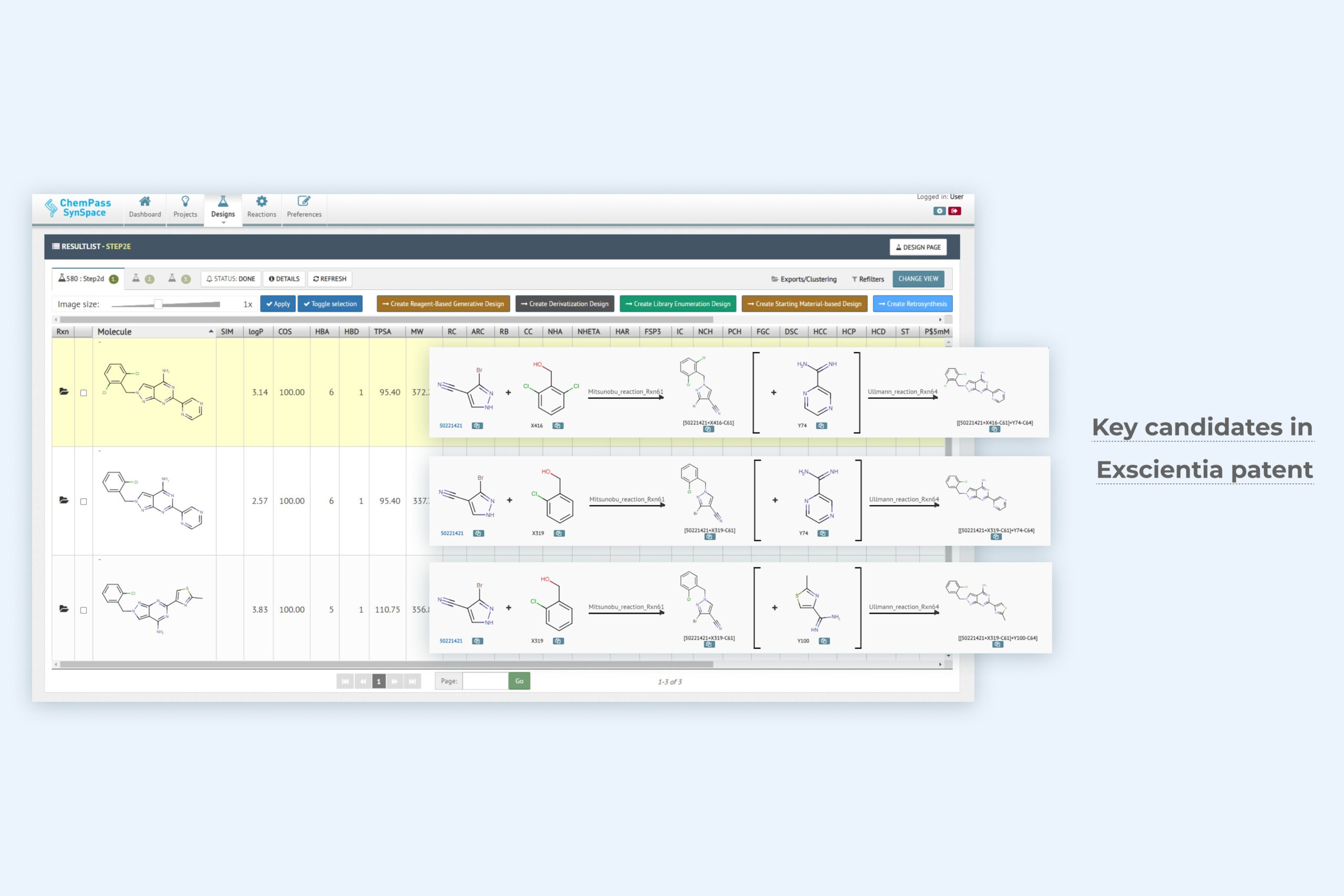
2. Library enumeration result page including three key analogs included in the final patent
Module returns a synthesizable focused library with all molecules that satisfy the enumeration criteria. Multiple positions can be varied in a single job. Synthesis cost and reagent availability can be used to rank close analogs.
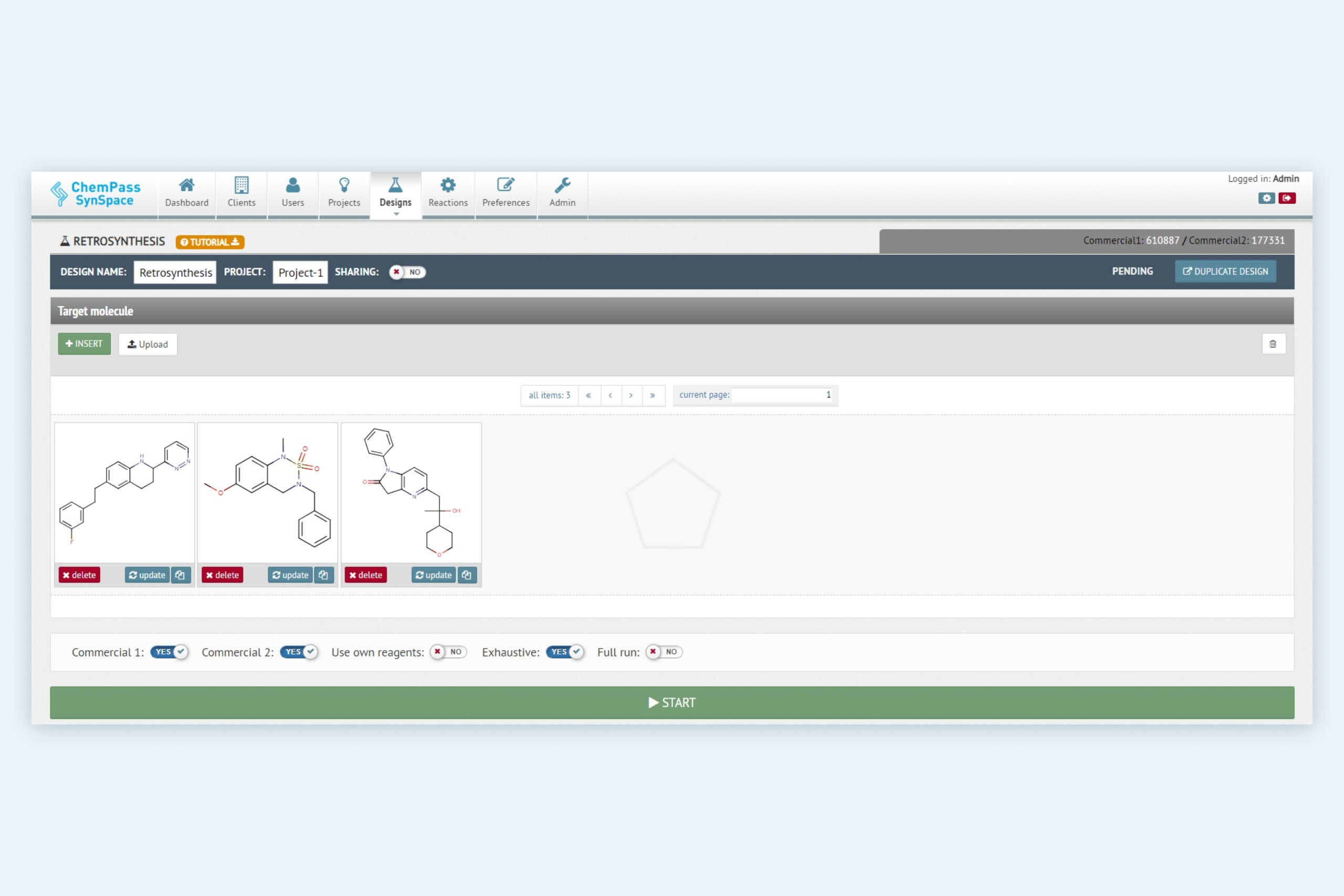
1. Retrosynthesis module setup page for retrosynthetic analysis and synthetic route ranking
Multiple molecules can be evaluated in a single job. The reagent sources can be customized. Can be run from GUI or via API.
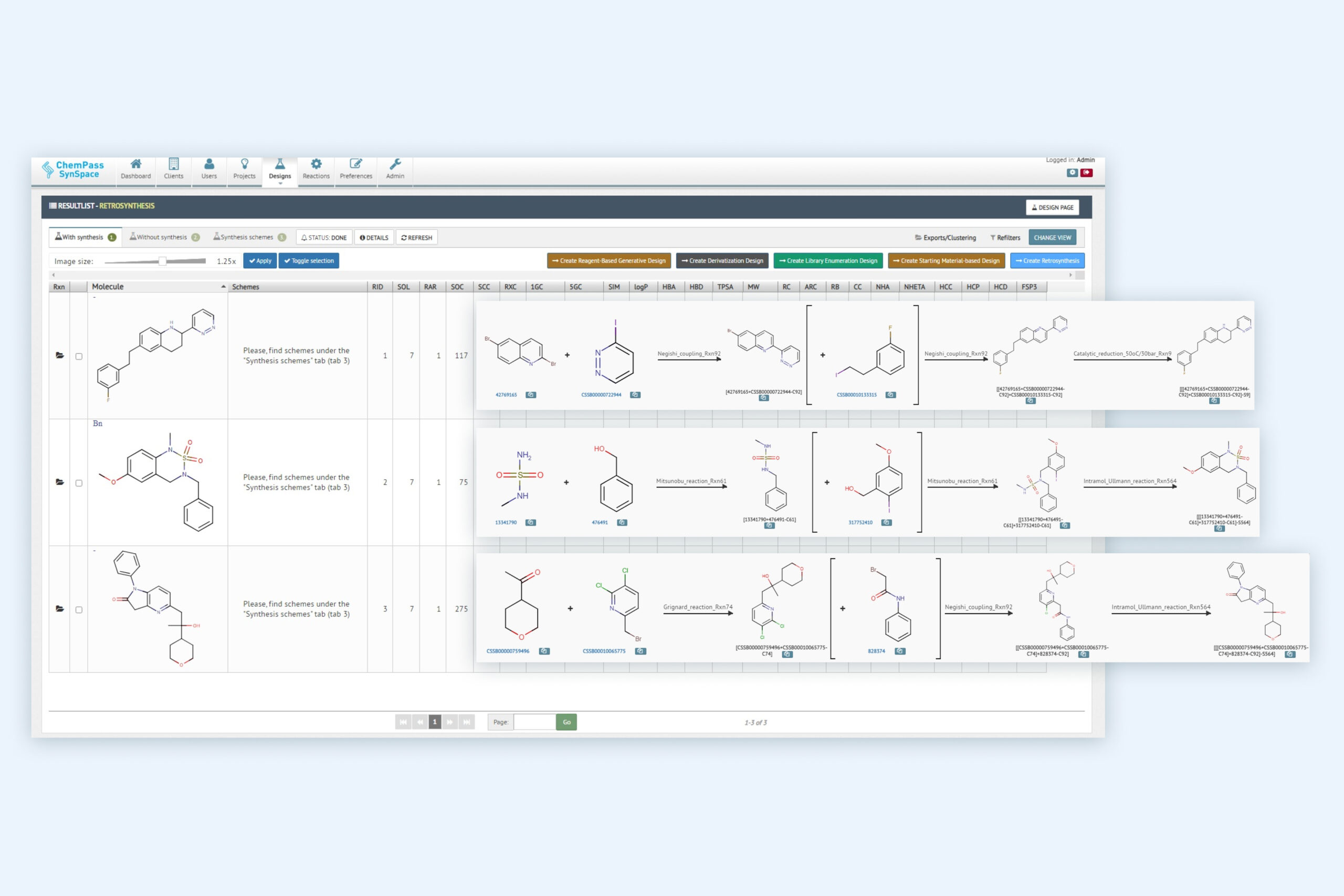
2. Retrosynthesis result page including multiple molecules with synthetic solutions
Module returns all molecules classified into solved and unsolved subsets. Solved molecules can contain up to 100,000 different routes with reagent cost and availability data as well as different ranking columns to assist selection of the best route(s) for synthesis or for focused library generation.